模型评估与优化常用方法实战
通过酶活性预测实战来体验体验模型从欠拟合到过拟合、再到拟合的过程。选择模型从线性回归 -> 多项式回归。通过进行异常检测,帮助找到了潜在的异常数据点,进行主成分分析,判断是否需要降低数据维度,然后通过数据分离,即使未提供测试样本,也能从训练数据中分离出测试数据。然后计算得到混淆矩阵,实现模型更全面的评估。最后通过调整KNN核心参数让模型在训练数据和测试数据上均得到了好的表现。
欠拟合与过拟合
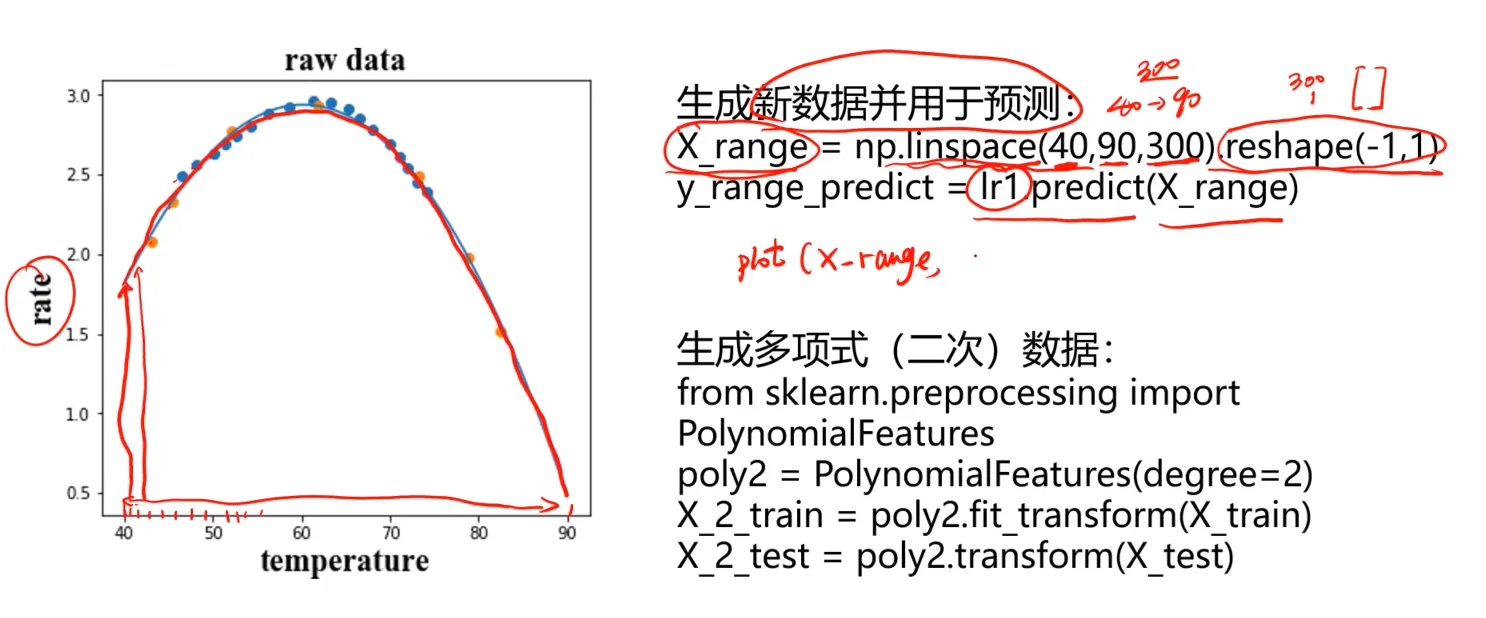 酶活性预测实战task:
酶活性预测实战task:
1、基于T-R-train.csv数据,建立线性回归模型,计算其在T-R-test.csv数据上的r2分数,可视化模型预测结果
2、加入多项式特征(2次、5次),建立回归模型
3、计算多项式回归模型对测试数据进行预测的r2分数,判断哪个模型预测更准确
4、可视化多项式回归模型数据预测结果,判断哪个模型预测更准确
下面直接贴Jupyter Notebook:
#load the data
import pandas as pd
import numpy as np
data_train = pd.read_csv('T-R-train.csv')
data_train.head()
| T | rate | |
|---|---|---|
| 0 | 46.53 | 2.49 |
| 1 | 48.14 | 2.56 |
| 2 | 50.15 | 2.63 |
| 3 | 51.36 | 2.69 |
| 4 | 52.57 | 2.74 |
#define X_train and y_train
X_train = data_train.loc[:,'T']
y_train = data_train.loc[:,'rate']
#visualize the data
from matplotlib import pyplot as plt
fig1 = plt.figure(figsize=(5,5))
plt.scatter(X_train,y_train)
plt.title('raw data')
plt.xlabel('temperature')
plt.ylabel('rate')
plt.show()
# 转换为N行1列的数据
X_train = np.array(X_train).reshape(-1,1)
先使用线行回归模型进行训练(其实通过看图其实我们已经大致能推断出线行模型不会有较好的表现):
#linear regression model prediction
from sklearn.linear_model import LinearRegression
lr1 = LinearRegression()
lr1.fit(X_train,y_train)
| LinearRegression() |
|---|
| on() |
#load the test data
data_test = pd.read_csv('T-R-test.csv')
X_test = data_test.loc[:,'T']
y_test = data_test.loc[:,'rate']
X_test = np.array(X_test).reshape(-1,1)
$$R^2=1-\frac{\sum_i(y_i-y_i)^2/n}{\sum_i(y_i-\hat{y})^2/n}=1-\frac{RMSE}{Var}$$
对于 $R^2$ 可以通俗地理解为使用均值作为误差基准,看预测误差是否大于或者小于均值基准误差。 R2_score = 1,样本中预测值和真实值完全相等,没有任何误差,表示回归分析中自变量对因变量的解释越好。 R2_score = 0。此时分子等于分母,样本的每项预测值都等于均值。
#make prediction on the training and testing data
y_train_predict = lr1.predict(X_train)
y_test_predict = lr1.predict(X_test)
from sklearn.metrics import r2_score
r2_train = r2_score(y_train,y_train_predict)
r2_test = r2_score(y_test,y_test_predict)
print('training r2:',r2_train)
print('test r2:',r2_test)
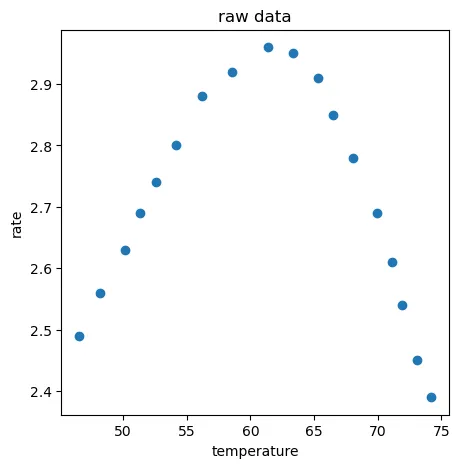
training r2: 0.016665703886981964
test r2: -0.758336343735132
因为图中温度大概是在40-90之间,所以生成300个40-90之间的X,也就是X_range
#generate new data 用于生成对应的预测值
X_range = np.linspace(40,90,300).reshape(-1,1)
y_range_predict = lr1.predict(X_range)
fig2 = plt.figure(figsize=(5,5))
plt.plot(X_range,y_range_predict)
plt.scatter(X_train,y_train)
plt.title('prediction data')
plt.xlabel('temperature')
plt.ylabel('rate')
plt.show()
很明显模型欠拟合,然后分别选择二次项多项式和五次项多项式模型进行回归:
#多项式模型
#generate new features
from sklearn.preprocessing import PolynomialFeatures
# 2阶多项式
poly2 = PolynomialFeatures(degree=2)
X_2_train = poly2.fit_transform(X_train)
X_2_test = poly2.transform(X_test)
# 5阶多项式
poly5 = PolynomialFeatures(degree=5)
X_5_train = poly5.fit_transform(X_train)
X_5_test = poly5.transform(X_test)
print(X_5_train.shape)
(18, 6)
lr2 = LinearRegression()
lr2.fit(X_2_train,y_train)
y_2_train_predict = lr2.predict(X_2_train)
y_2_test_predict = lr2.predict(X_2_test)
r2_2_train = r2_score(y_train,y_2_train_predict)
r2_2_test = r2_score(y_test,y_2_test_predict)
lr5 = LinearRegression()
lr5.fit(X_5_train,y_train)
y_5_train_predict = lr5.predict(X_5_train)
y_5_test_predict = lr5.predict(X_5_test)
r2_5_train = r2_score(y_train,y_5_train_predict)
r2_5_test = r2_score(y_test,y_5_test_predict)
print('training r2_2:',r2_2_train)
print('test r2_2:',r2_2_test)
print('training r2_5:',r2_5_train)
print('test r2_5:',r2_5_test)
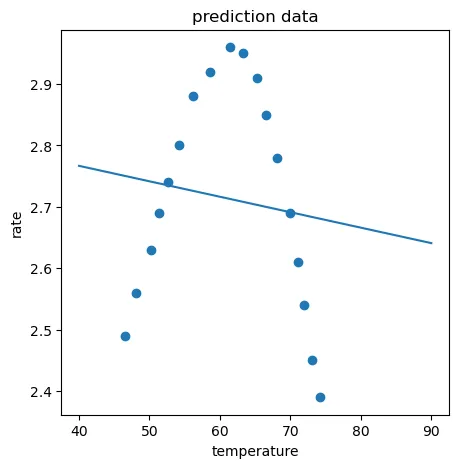
training r2_2: 0.9700515400689426
test r2_2: 0.9963954556468683
training r2_5: 0.9978527267327939
test r2_5: 0.5437885877449662
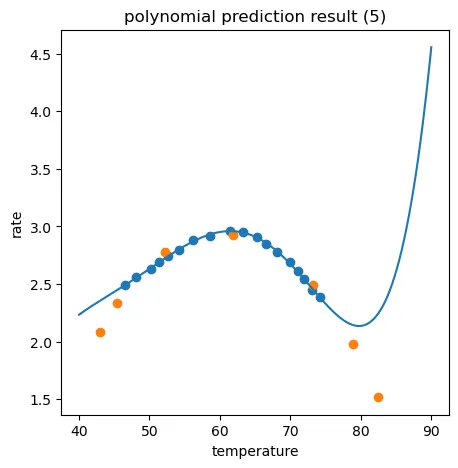
二次项回归模型很明显是比较合适的,五次项回归虽然对于训练数据有0.9978的分数,但是对于测试数据却差强人意,这就是明显的过拟合。
X_2_range = np.linspace(40,90,300).reshape(-1,1)
X_2_range = poly2.transform(X_2_range)
y_2_range_predict = lr2.predict(X_2_range)
X_5_range = np.linspace(40,90,300).reshape(-1,1)
X_5_range = poly5.transform(X_5_range)
y_5_range_predict = lr5.predict(X_5_range)
同样通过生成数据的方式分别看看二次项回归模型(正好拟合)和五次项回归模型(过拟合)的图形化展示:
fig3 = plt.figure(figsize=(5,5))
plt.plot(X_range,y_2_range_predict)
plt.scatter(X_train,y_train)
plt.scatter(X_test,y_test)
plt.title('polynomial prediction result (2)')
plt.xlabel('temperature')
plt.ylabel('rate')
plt.show()
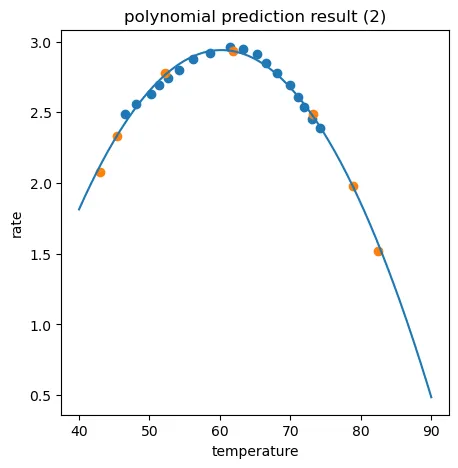
fig4 = plt.figure(figsize=(5,5))
plt.plot(X_range,y_5_range_predict)
plt.scatter(X_train,y_train)
plt.scatter(X_test,y_test)
plt.title('polynomial prediction result (5)')
plt.xlabel('temperature')
plt.ylabel('rate')
plt.show()
酶活性预测实战:
1、通过建立二阶多项式回归模型,对酶活性实现了一个较好的预测,无论针对训练或测试数据都得到一个高的r2分数;
2、通过建立线性回归、五阶多项式回归模型,发现存在过拟合或欠拟合情况。过拟合情况下,对于训练数据r2分数高(预测准确),但对于预测数据r2分数低(预测不准确);
3、无论时通过r2分数,或是可视化模型结果,都可以发现二阶多项式回归模型效果最好;
4、核心算法参考链接: https://scikit-learn.org/stable/modules/generated/…
高斯分布去除异常点
基于data_class_raw.csv数据,根据高斯分布概率密度函数,寻找异常点并剔除
#load the data
import pandas as pd
import numpy as np
data = pd.read_csv('data_class_raw.csv')
data.head()
| x1 | x2 | y | |
|---|---|---|---|
| 0 | 0.77 | 3.97 | 0 |
| 1 | 1.71 | 2.81 | 0 |
| 2 | 2.18 | 1.31 | 0 |
| 3 | 3.80 | 0.69 | 0 |
| 4 | 5.21 | 1.14 | 0 |
#define X and y
X = data.drop(['y'],axis=1)
y = data.loc[:,'y']
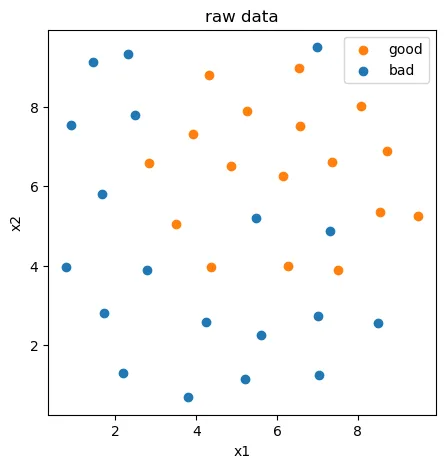
#visualize the data
from matplotlib import pyplot as plt
fig1 = plt.figure(figsize=(5,5))
bad = plt.scatter(X.loc[:,'x1'][y==0],X.loc[:,'x2'][y==0])
good = plt.scatter(X.loc[:,'x1'][y==1],X.loc[:,'x2'][y==1])
plt.legend((good,bad),('good','bad'))
plt.title('raw data')
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
from sklearn.covariance import EllipticEnvelope
ad_model = EllipticEnvelope(contamination=0.02)
ad_model.fit(X[y==0])
y_predict_bad = ad_model.predict(X[y==0])
print(y_predict_bad)
[ 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 -1]
fig2 = plt.figure(figsize=(5,5))
bad = plt.scatter(X.loc[:,'x1'][y==0],X.loc[:,'x2'][y==0])
good = plt.scatter(X.loc[:,'x1'][y==1],X.loc[:,'x2'][y==1])
plt.scatter(X.loc[:,'x1'][y==0][y_predict_bad==-1],X.loc[:,'x2'][y==0][y_predict_bad==-1], marker='x',s=150)
plt.legend((good,bad),('good','bad'))
plt.title('raw data')
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
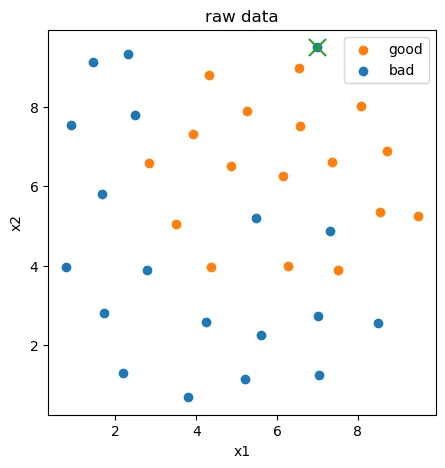
此时不管是用代码删除也好,还是手动删除csv的数据也行,只要去掉异常数据点即可。
PCA分析与降维
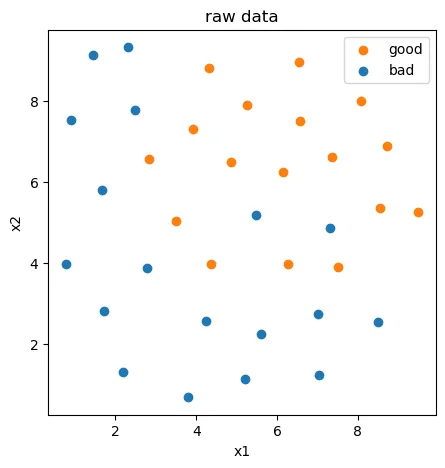
基于data_class_processed.csv数据(也就是去掉异常点之后的数据),进行PCA处理,确定重要数据维度及成分。
# 此时操作就是已经经过异常处理之后的数据
data = pd.read_csv('data_class_processed.csv')
data.head()
#define X and y
X = data.drop(['y'],axis=1)
y = data.loc[:,'y']
fig3 = plt.figure(figsize=(5,5))
bad = plt.scatter(X.loc[:,'x1'][y==0],X.loc[:,'x2'][y==0])
good = plt.scatter(X.loc[:,'x1'][y==1],X.loc[:,'x2'][y==1])
plt.legend((good,bad),('good','bad'))
plt.title('raw data')
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
# pca
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
# 标准化处理
X_nomr = StandardScaler().fit_transform(X)
pca = PCA(n_components=2)
X_reduced = pca.fit_transform(X_nomr)
# 计算维度的标准差比例
var_ratio = pca.explained_variance_ratio_
print(var_ratio)
[0.5369408 0.4630592]
fig4 = plt.figure(figsize=(5,5))
plt.bar([1,2],var_ratio)
plt.show()
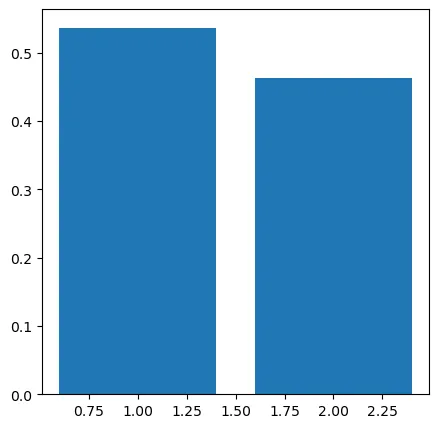
经过主成分分析,可以看出,这两个维度的都是需要保留的。
训练与测试数据分离
# 数据分离
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,random_state=4,test_size=0.4)
print(X_train.shape, X_test.shape, X.shape)
(21, 2) (14, 2) (35, 2)
建立KNN模型
建立KNN模型,完成分类任务
from sklearn.neighbors import KNeighborsClassifier
knn_10 = KNeighborsClassifier(n_neighbors=10)
knn_10.fit(X_train,y_train)
y_train_predict = knn_10.predict(X_train)
y_test_predict = knn_10.predict(X_test)
# 看下准确率
from sklearn.metrics import accuracy_score
train_acc = accuracy_score(y_train,y_train_predict)
test_acc = accuracy_score(y_test,y_test_predict)
print(train_acc, test_acc)
0.9047619047619048 0.6428571428571429
测试数据表现不太理想
#可视化分类边界
xx,yy = np.meshgrid(np.arange(0,10,0.05), np.arange(0,10,0.05))
print(xx.shape, yy.shape)
(200, 200) (200, 200)
x_range = np.c_[xx.ravel(),yy.ravel()]
print(x_range.shape)
(40000, 2)
y_range_predict = knn_10.predict(x_range)
fig5 = plt.figure(figsize=(5,5))
knn_bad = plt.scatter(x_range[:,0][y_range_predict==0],x_range[:,1][y_range_predict==0])
knn_good = plt.scatter(x_range[:,0][y_range_predict==1],x_range[:,1][y_range_predict==1])
bad = plt.scatter(X.loc[:,'x1'][y==0],X.loc[:,'x2'][y==0])
good = plt.scatter(X.loc[:,'x1'][y==1],X.loc[:,'x2'][y==1])
plt.legend((good,bad,knn_good,knn_bad),('good','bad','knn_good','knn_bad'))
plt.title('predict_result')
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
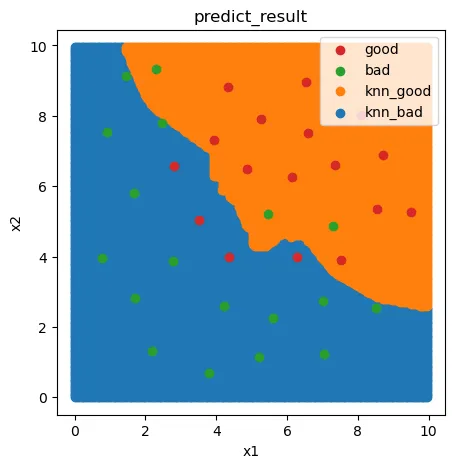
混淆矩阵评估
# 计算混淆矩阵
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_test_predict)
print(cm)
[[4 2]
[3 5]]
TP = cm[1,1]
TN = cm[0,0]
FP = cm[0,1]
FN = cm[1,0]
print(TP,TN,FP,FN)
5 4 2 3
#准确率: 整体样本中,预测正确样本数的比例
accuracy = (TP + TN)/(TP + TN + FP + FN)
print(accuracy)
0.6428571428571429
#灵敏度(召回率): 正样本中,预测正确的比例
recall = TP/(TP + FN)
print(recall)
0.625
#特异度: 负样本中,预测正确的比例
specificity = TN/(TN + FP)
print(specificity)
0.6666666666666666
#精确率: 预测结果为正的样本中,预测正确的比例
precision = TP/(TP + FP)
print(precision)
0.7142857142857143
#F1分数: 综合Precision和Recall的一个判断指标
#F1 Score = 2*Precision X Recall/(Precision + Recall)
f1 = 2*precision*recall/(precision+recall)
print(f1)
0.6666666666666666
调整至合适参数
#try different k and calcualte the accuracy for each
n = [i for i in range(1,21)]
accuracy_train = []
accuracy_test = []
for i in n:
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(X_train,y_train)
y_train_predict = knn.predict(X_train)
y_test_predict = knn.predict(X_test)
accuracy_train_i = accuracy_score(y_train,y_train_predict)
accuracy_test_i = accuracy_score(y_test,y_test_predict)
accuracy_train.append(accuracy_train_i)
accuracy_test.append(accuracy_test_i)
print(accuracy_train,accuracy_test)
[1.0, 1.0, 1.0, 1.0, 1.0, 0.9523809523809523, 0.9523809523809523, 0.9523809523809523, 0.9047619047619048, 0.9047619047619048, 0.9047619047619048, 0.9523809523809523, 0.9047619047619048, 0.9047619047619048, 0.9523809523809523, 0.9047619047619048, 0.9047619047619048, 0.5714285714285714, 0.5714285714285714, 0.5714285714285714] [0.5714285714285714, 0.5, 0.5, 0.5714285714285714, 0.7142857142857143, 0.5714285714285714, 0.5714285714285714, 0.5714285714285714, 0.6428571428571429, 0.6428571428571429, 0.6428571428571429, 0.5714285714285714, 0.6428571428571429, 0.6428571428571429, 0.5714285714285714, 0.5714285714285714, 0.5714285714285714, 0.42857142857142855, 0.42857142857142855, 0.42857142857142855]
fig6 = plt.figure(figsize=(12,5))
plt.subplot(121)
plt.plot(n,accuracy_train,marker='o')
plt.title('training accuracy vs n_neighbors')
plt.xlabel('n_neighbors')
plt.ylabel('accuracy')
plt.subplot(122)
plt.plot(n,accuracy_test,marker='o')
plt.title('testing accuracy vs n_neighbors')
plt.xlabel('n_neighbors')
plt.ylabel('accuracy')
plt.show()
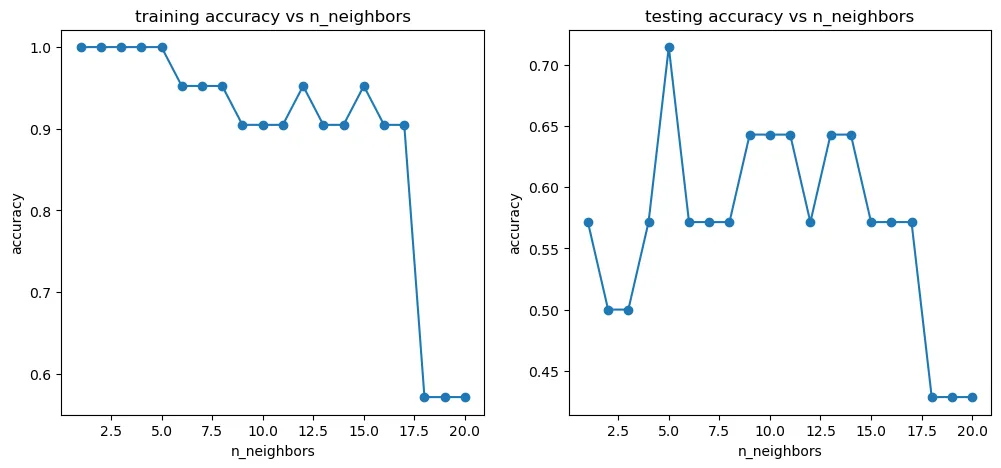
可以看出,n_neighbors 参数大约在5的时候,不管是对于训练数据还是测试数据,表现都还是不错的
from sklearn.neighbors import KNeighborsClassifier
knn_5 = KNeighborsClassifier(n_neighbors=5)
knn_5.fit(X_train,y_train)
y_train_predict = knn_5.predict(X_train)
y_test_predict = knn_5.predict(X_test)
# 看下准确率
from sklearn.metrics import accuracy_score
train_acc = accuracy_score(y_train,y_train_predict)
test_acc = accuracy_score(y_test,y_test_predict)
print(train_acc, test_acc)
1.0 0.7142857142857143
重新绘制一下分类边界:
y_range_predict = knn_5.predict(x_range)
fig6 = plt.figure(figsize=(5,5))
knn_bad = plt.scatter(x_range[:,0][y_range_predict==0],x_range[:,1][y_range_predict==0])
knn_good = plt.scatter(x_range[:,0][y_range_predict==1],x_range[:,1][y_range_predict==1])
bad = plt.scatter(X.loc[:,'x1'][y==0],X.loc[:,'x2'][y==0])
good = plt.scatter(X.loc[:,'x1'][y==1],X.loc[:,'x2'][y==1])
plt.legend((good,bad,knn_good,knn_bad),('good','bad','knn_good','knn_bad'))
plt.title('when n=5 predict_result')
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
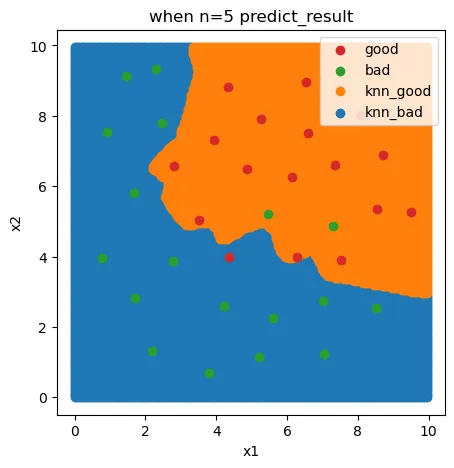
好坏质检分类实战summary:
1、通过进行异常检测,帮助找到了潜在的异常数据点;
2、通过PCA分析,发现需要保留2维数据集;
3、实现了训练数据与测试数据的分离,并计算模型对于测试数据的预测准确率
4、计算得到混淆矩阵,实现模型更全面的评估
5、通过新的方法,可视化分类的决策边界
6、通过调整核心参数n_neighbors值,在计算对应的准确率,可以帮助我们更好的确定使用哪个模型
7、核心算法参考链接: https://scikit-learn.org/stable/modules/generated/…